Summary #
Cardiovascular diseases cause over 18 million deaths globally every year, occurring mostly as myocardial infarctions and heart failures. This disease has globally accounted for about 33% of all causes of deaths in 2019 alone. Heart failure occurs when the heart is not capable of pumping enough blood to meet the body’s needs. Machine learning has the ability to predict the most important features from the patients’ medical records that are responsible for heart failure. The goal of this analysis is to confirm that serum creatinine blood levels and heart ejection fraction, which is the percentage of blood leaving the heart at each contraction, are the two most relevant features responsible for predicting heart failure in affected patients, as stated in the main research article used for this analysis.
Introduction #
The Heart failure clinical records dataset, collected in 2015, contains the medical records of 299 patients who had heart failure. The dataset was collected during their follow-up period, and contains 13 clinical features. These features, both numerical and categorical, are patient age, presence or absence of anaemia, level of creatinine phosphokinase in the blood, presence or absence of diabetes, ejection fraction, presence or absence of high blood pressure, platelet level in the blood, level of serum creatinine in the blood, level of serum sodium in the blood, patient sex, presence or absence of patient smoking, follow-up check-up time, and death event. This is a classification problem, with the death event column as the binary outcome column. The research article can be accessed at the link, Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone (link to pdf here ). The data set can be retrieved from the UCI Machine Learning Repository at the link, Heart failure clinical records .
Exploratory Data Analysis #
Initially from the exploratory data analysis, aside from the evident dependence of heart failure on age itself, one may be able to glimpse at a dependence of fatal heart failures from the serum creatinine and serum sodium blood levels, and from the ejection fraction, which is defined as the percentage of blood leaving the heart at each contraction (see Figure 1 below). Although interesting in itself, further analyses from the exploratory data analysis observations are still warranted to obtain a complete picture of the data set.
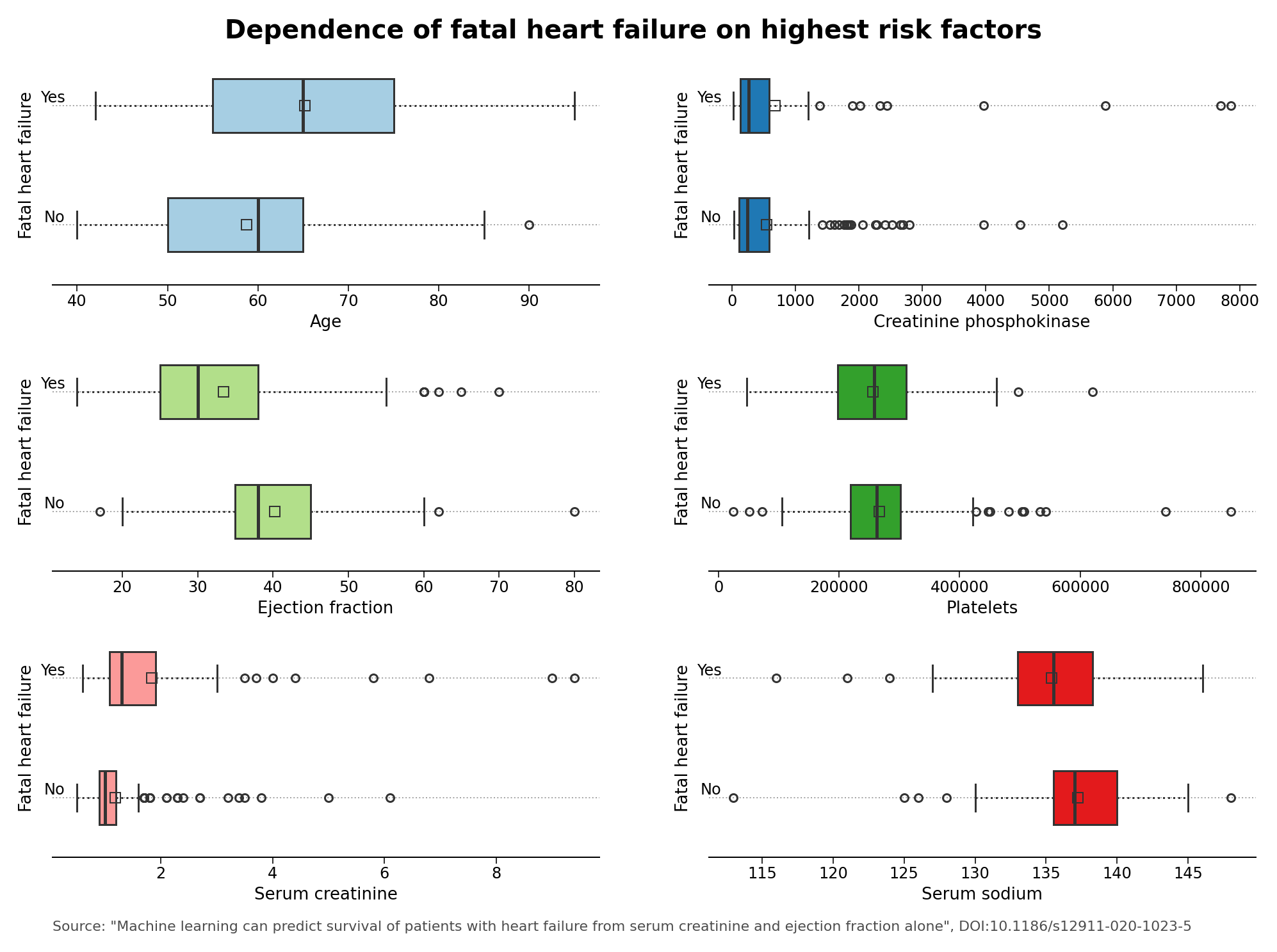
Figure 1 – Dependence of heart failure on highest risk factors

Model Analysis #
The research article based on the data set states that a random forest model is the best model that can predict survival of patients with heart failure from the serum creatinine and ejection fraction features alone. To verify this result, the model analysis was performed using several different machine learning algorithms, such as LogisticRegression , DecisionTreesClassifier , RandomForestsClassifier from Scikit-Learn and XGBoostClassifier from XGBoost . The models were trained on a training set, representing 60% of the data, and evaluated on a validation set, representing 20% of the data. The categorical features in the dataset were transformed using the DictVectorizer class from Scikit-Learn. A table of values of the F1 score and Area Under the Curve (AUC) values was generated for all of the classifiers (Table 1).
| Classifier | F1 score | AUC |
|---|---|---|
| Logistic Regression | 0.727 | 0.791 |
| Decision Trees | 0.737 | 0.807 |
| eXtreme Gradient Boost (XGBoost) | 0.743 | 0.806 |
| Random Forests | 0.778 | 0.832 |
Hyperparameter tuning on the Random Forests model was undertaken using the GridSearchCV class from Scikit-Learn. Due to the low count of entries in the dataset (299), the hyperparameter tuning did not achieve a better result in F1 score and AUC value compared to the default hyperparameters, and hence the hypertuned parameters were discarded. Using the combined training and validation sets to generate the Random Forest model, the values obtained for F1 score and AUC value resulted in being very similar to the previous values for the same classifier, as shown in the Table 2.
| Classifier | F1 score | AUC |
|---|---|---|
| Random Forests | 0.769 | 0.834 |
The feature importance plot was generated using the default hyperparameters values of the Random Forests model. The feature importance plot shows that the features of greatest importance after follow-up check-up time are indeed serum creatinine and ejection fraction, as stated in the main research article of the data set.
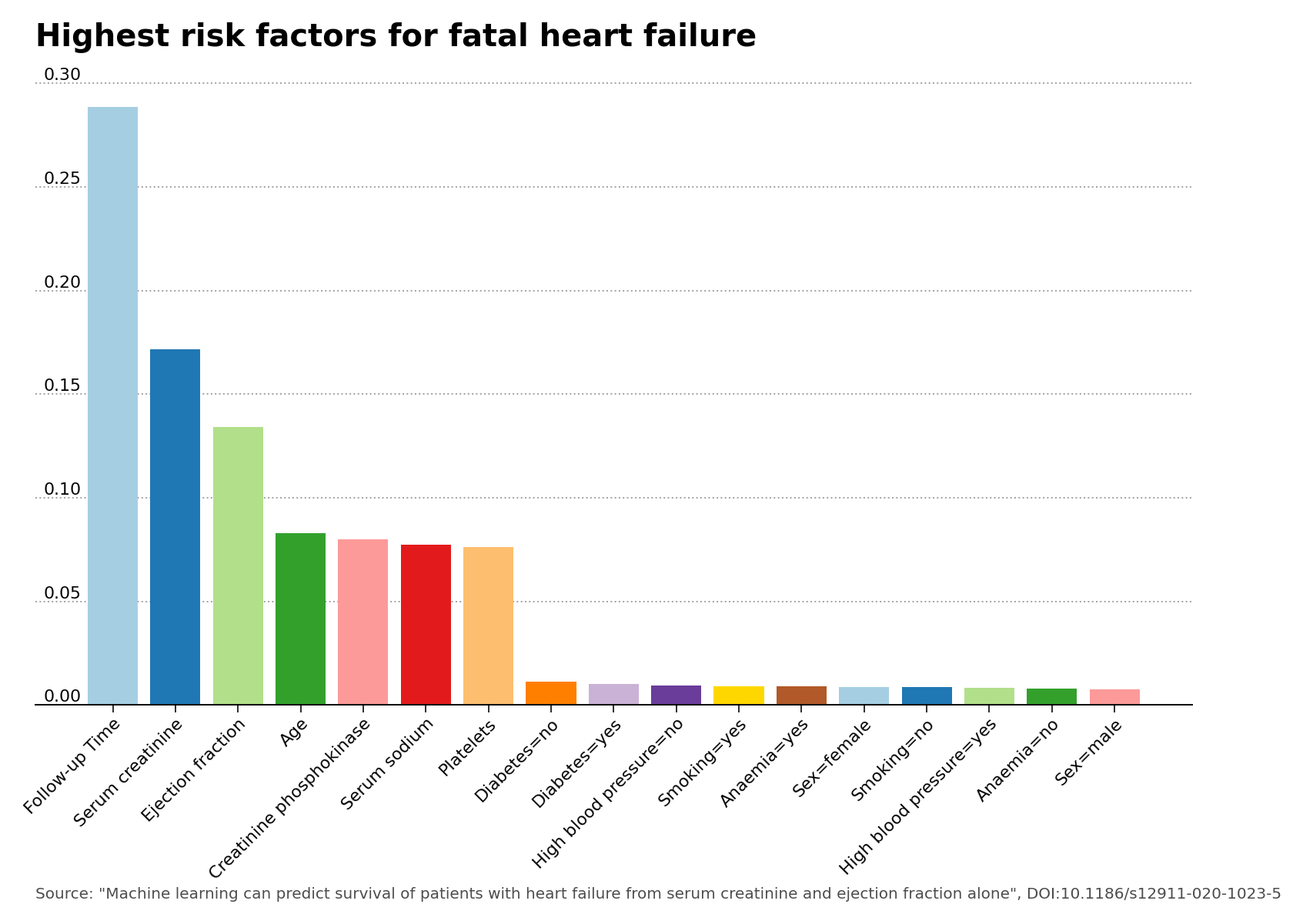
Figure 2 – Highest risk factors for fatal heart failure.

Deployment With Docker #
The Random Forest model was subsequently stored in a Python environment using Pipenv
, containerized using a Docker
container, and then was deployed online on Render
. By using a Python script from my GitHub project repository, the Docker container can return predictions that lead to a positive and a negative case of heart failure, from two pre-defined sets of patient characteristics, obtained from their medical records, that are included in the script. The Docker container was first tested locally before being uploaded remotely on Docker Hub and then Render. There is also a Python script to locally test the container using the same patient data.
The details to obtain the predictions are listed in the description section of the project on my GitHub repository. The description section also gives a detailed list of the use of each file. It also describes how to build the Python environment and the Docker container that contains the model.
Conclusions #
The best model proved to be the Random Forests classifier, and due to the low amount of entries in the dataset, no improvement was gained by performing hyperparameter tuning. The feature importance plot did indeed show that the serum creatinine and ejection fraction features can be predictors of the survival of patients with heart failure. The model is made available online as a Docker container, and can deliver heart failure predictions by use of a specific Python script.
Feel free to take a look at the code of my project in my GitHub repository. For the data analysis, the following software packages were used: Scikit-Learn (version 1.2.2), pandas (version 2.1.4), matplotlib (version 3.8.0) and Docker Desktop on Mac (4.26.1, build 131620).
